Central to all life are diverse fundamental genetic processes. It is thus crucial to have a detailed understanding of genetic concepts and the molecular mechanisms that determine traits in organisms. Genetic defects and dysfunctions in genetic processes are the major players in the development of complex conditions, such as cancer, cardiovascular, metabolic, and neurodegenerative diseases. Increasingly, applications of genetic discoveries are crossing over into more and different areas of society such as agriculture, medicine, and forensics; it is therefore essential to further our knowledge and promote genetics literacy to the societal and health care setting as a whole. A firm understanding of genetic processes offers further the promise of novel gene therapy approaches to treat diseases that are currently not treatable otherwise.
CHROMOSOME ORGANIZATION AND SEGREGATION
⦿ all eukyryotic SMCs induce negative twist during each loop extrusion step

Eukaryotes carry three types of Structural Maintenance of Chromosomes (SMC) protein complexes, condensin, cohesin, and SMC5/6, which are ATP-dependent motor proteins that remodel the genome via DNA loop extrusion. SMCs modulate DNA supercoiling, but it has remained incompletely understood how this is achieved. Here we present a single-molecule magnetic tweezers assay that directly measures how much twist is induced by an individual SMC in each loop-extrusion step. We demonstrate that all three SMC complexes induce the same large negative twist (i.e., a linking number change ΔLk of -0.6 at each loop-extrusion step) into the extruded loop. This -0.6 twist is found to be independent of step size. Using ATP-hydrolysis mutants and non-hydrolysable ATP analogues, we find that ATP binding is the twist-inducing event during the ATPase cycle, which coincides with the force-generating loop-extrusion step. The fact that all three eukaryotic SMC proteins induce the same amount of twist indicates a similar DNA-loop-extrusion mechanism among these SMC complexes.
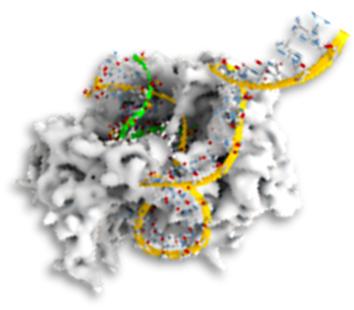
⦿ ParB:DNA interactions create partition condensates in bacterial chromosome segregation

In most bacteria, chromosome segregation is driven by the ParABS system where the CTPase protein ParB loads at the parS site to trigger the formation of a large partition complex. Here, we present in vitro studies of the partition complex for Bacillus subtilis ParB, using single- molecule fluorescence microscopy and AFM imaging to show that transient ParB-ParB bridges are essential for forming DNA condensates. Molecular Dynamics simulations confirm that condensation occurs abruptly at a critical concentration of ParB and show that multimerization is a prerequisite for forming the partition complex. Magnetic tweezer force spectroscopy on mutant ParB proteins demonstrates that CTP hydrolysis at the N-terminal domain is essential for DNA condensation. Finally, we show that transcribing RNA polymerases can steadily traverse the ParB-DNA partition complex. These findings uncover how ParB forms a stable yet dynamic partition complex for chromosome segregation that induces DNA condensation and segregation while enabling replication and transcription. >Download Study<
⦿ CTCF is a DNA-tension-dependent barrier to cohesin-mediated DNA loop extrusion

In eukaryotes, genomic DNA is extruded into loops by cohesin. By restraining this process, the DNA-binding protein CCCTC-binding factor (CTCF) generates topologically associating domains (TADs) that have important roles in gene regulation and recombination during development and disease. How CTCF establishes TAD boundaries and to what extent these are permeable to cohesin is unclear. Here, to address these questions, we visualize interactions of single CTCF and cohesin molecules on DNA in vitro. We show that CTCF is sufficient to block diffusing cohesin, possibly reflecting how cohesive cohesin accumulates at TAD boundaries, and is also sufficient to block loop-extruding cohesin, reflecting how CTCF establishes TAD boundaries. CTCF functions asymmetrically, as predicted; however, CTCF is dependent on DNA tension. Moreover, CTCF regulates cohesin’s loop-extrusion activity by changing its direction and by inducing loop shrinkage. Our data indicate that CTCF is not, as previously assumed, simply a barrier to cohesin-mediated loop extrusion but is an active regulator of this process, whereby the permeability of TAD boundaries can be modulated by DNA tension. These results reveal mechanistic principles of how CTCF controls loop extrusion and genome architecture. >Download Study<
⦿ DNA flexibility governs Condensin-extruded DNA loop length

The condensin SMC protein complex organizes chromosomal structure by extruding loops of DNA. Its ATP-dependent motor mechanism remains unclear but likely involves steps associated with large conformational changes within the ∼50 nm protein complex. Here, using high-resolution magnetic tweezers, we resolve single steps in the loop extrusion process by individual yeast condensins. The measured median step sizes range between 20–40 nm at forces of 1.0–0.2 pN, respectively, comparable with the holocomplex size. These large steps show that, strikingly, condensin typically reels in DNA in very sizeable amounts with ∼200 bp on average per single extrusion step at low force, and occasionally even much larger, exceeding 500 bp per step. Using Molecular Dynamics simulations, we demonstrate that this is due to the structural flexibility of the DNA polymer at these low forces. Using ATP-binding-impaired and ATP-hydrolysis-deficient mutants, we find that ATP binding is the primary step-generating stage underlying DNA loop extrusion. We discuss our findings in terms of a scrunching model where a stepwise DNA loop extrusion is generated by an ATP-binding-induced engagement of the hinge and the globular domain of the SMC complex. >Download Study<
⦿ Chromosome Compaction in Stationary Bacteria Does Not Affect gene expression

This work work provides a surprising new chapter to a long and evolving debate on the regulation of transcription in prokaryotes. The currently accepted consensus describes that prokaryotes, similar to eukaryotes, employ compaction of the genome as a mechanism to repress transcription via nucleoid associated proteins (NAPs). We investigated the capacity of genome compaction and impact on protein biogenesis of the protein Dps, a NAP that is the predominant component of the nucleoid during starvation. Our findings challenge the existing consensus that nucleoid architecture is intimately connected to transcription: while the expression of Dps induces a fundamental change in the nucleoid architecture, we found no significant effect of Dps on gene expression. We further observed that while the RNA polymerase and transcription factors had full access to the compacted genome, restriction enzymes were blocked. We conclude that Dps has evolved to act orthogonally to transcription. Our results are the first direct evidence that a nucleoid-compacting NAP does not affect biogenesis and that phase-separated organelles are employed in prokaryotes. >Download Study<
⦿ EPIGENETICS: INHIBITION OF MLL1 RECRUITMENT TO CHROMATIN AS NOVEL THERAPEUTIC TARGET

CpG islands recruit MLL1 via the CXXC domain to modulate chromatin structure and regulate gene expression. The amino acid motif of CXXC also plays a pivotal role in MLL1’s structure and function and serves as a target for drug design. In addition, the CpG pattern in an island governs spatially dependent collaboration among CpGs in recruiting epigenetic enzymes. However, current studies using short DNA fragments cannot probe the dynamics of CXXC on long DNA with crowded CpG motifs. Here, we used single-molecule magnetic tweezers to examine the binding dynamics of MLL1’s CXXC domain on a long DNA with a CpG island. The mechanical strand separation assay allows profiling of protein-DNA complexes and reports force-dependent unfolding times. Further design of a hairpin detector reveals the unfolding time of individual CXXC-CpG complexes. Finally, in a proof of concept we demonstrate the inhibiting effect of dimethyl fumarate on the CXXC-DNA complexes by measuring the dose response curve of the unfolding time. This demonstrates the potential feasibility of using single-molecule strand separation as a label-free detector in drug discovery and chemical biology. >Download Study<
TRANSCRIPTION, DNA REPLICATION & RECOMBINATION
⦿ Supercoil induction during transcription is independent from drag of RNA transcript

Transcription-coupled supercoiling of DNA is key in chromosome compaction and the regulation of genetic processes in all domains of life. It has become common knowledge that, during transcription, the DNA-dependent RNA polymerase (RNAP) induces positive supercoiling downstream and negative supercoils upstream, as rotation of RNAP around the DNA axis gets constrained due to drag on its RNA transcript. Here, we experimentally validate this so-called twin-supercoiled-domain model in vitro at real-time at the single-molecule scale. Upon binding to the promoter site on a supercoiled DNA molecule, RNAP merges all DNA supercoils into one large pinned plectoneme with RNAP residing at its apex. Transcription by RNAP in real time demonstrates that up- and downstream supercoils are generated simultaneously and in equal portions, in agreement with the twin-supercoiled-domain model. Experiments carried out in the presence of RNases A and H, revealed that an additional viscous drag of the RNA transcript is not necessary for the RNAP to induce supercoils. The latter results contrast the current consensus and simulations on the origin of the twin-supercoiled domains, pointing at an additional mechanistic cause underlying supercoil generation by RNAP. >Download Study<
⦿ discovered mechanism of recombination in RNA viruses as novel therapeutic target

RNA viruses pose a serious threat to global public health. Genetic recombination is a crucial step in many RNA viruses, contributing to viral diversity and chimeric subspecies, which regularly causes new outbreaks. This process challenges the development of vaccines and antiviral medication. A detailed understanding of the mechanism and triggers underlying recombination remains to be identified and could provide new ideas for antiviral therapies. To elucidate the mechanism of recombination and the effect of a new class of antiviral agents, we used a unique ensemble of in vivo and in vitro methods to probe Enterovirus A71 and Poliovirus. We show that recombination is an RdRp-mediated process and that virus sequence impacts recombination efficiency and virulence in vivo. In contrast, we observed at the single-molecule level that recombination depends on the extent of RdRp pausing and temporal arrest. Importantly, recombination in both RNA viruses is increased by antiviral agents that induce a significant RdRp arrest. Our results indicate a conserved recombination mechanism across RNA viruses, which further represents a novel target for antiviral therapy with broad-spectrum appeal. >Download Study< Published method protocol here: >Download Protocol<
⦿ Transcription: a unifying model of 3 pause states and non-diffusive backtrack recovery

RNA polymerases (RNAp) are the main target for the regulation of gene expression. Pausing by RNAp facilitates the recruitment of regulatory factors, RNA folding, and other related processes. While backtracking and intra-structural isomerization have been proposed to trigger pausing, the mechanisms of pause formation and recovery remain debated. Using high-throughput magnetic tweezers with single-molecule sensitivity, we have examined the full temporal spectrum of Escherichia coli RNAp transcription dynamics. Together with probabilistic dwell-time analysis and modelling, this has identified three distinct states that compete with elongation: a short-lived elemental pause and two long-lived backtracked pause states. We demonstrate that recovery from backtracking is not governed by diffusional Brownian motion, but rather by intrinsic RNA cleavage, whereby RNAp conformational changes hinder cleavage in long-lived pause states. We further show that state switching underlies stochastic alterations in the frequency of short pauses. Based on these findings, we propose a consensus model of intrinsic pausing that unifies all key findings while resolving earlier contradictions. >Download Study<
⦿ A tool to Predicting Intraserotypic Recombination in Enterovirus 71

Enterovirus 71 (EV-A71), related to the poliovirus (PV) model, causes similar clinical outcomes of acute flaccid paralysis in recurrent outbreaks throughout Asia. Phylogenetic analysis has shown that recombination between co-circulating strains of EV-A71 produces the outbreak-associated strains with increased virulence and transmissibility. To date, still little is known about factors that influence recombination in EV-A71. We developed a cell-based assay to study recombination of EVA71 and show that: (1) EV-A71 strain-type and RNA sequence diversity impacts recombination frequency in a predictable manner that mimics the observations found in nature; (2) recombination is primarily a replicative process mediated by the RNA-dependent RNA polymerase (RdRp); (3) a mutation shown to reduce recombination in PV similarly reduces EV-A71 recombination suggesting conservation in mechanism(s); (4) sequencing of recombinant genomes indicates that template switching mechanism requires sequence homology at the recombination junction while triggers for template-switching is sequence-independent. This recombination assay will allow investigating the interplay between replication, recombination and disease. >Download Study<
⦿ DNA Replication: DNA strand separation induces lock at the Tus-Ter replication fork

The bidirectional replication of a circular chromosome by many bacteria necessitates proper termination to avoid the head-on collision of the opposing replisomes during DNA replication. In E. Coli, replisome progression beyond the termination site is prevented by Tus proteins bound to asymmetric Ter sites. Previous stidies indicate that strand separation on the blocking (non-permissive) side of Tus–Ter triggers roadblock formation, but biochemical evidence also suggests roles for protein-protein interactions. Here, single-molecule DNA unzipping experiments demonstrate that non-permissively oriented Tus–Ter forms a tight lock in the absence of replicative proteins, whereas permissively oriented Tus–Ter allows nearly unhindered strand separation. Quantifying the lock strength revealed the existence of several intermediate lock states that are impacted by mutations in the lock domain, but not by mutations in the DNA-binding domain. Our results demonstrate that the lock formation is highly specific and exceeds reported in vivo efficiencies. We postulate that protein-protein interactions may actually hinder, rather than promote, proper lock formation. >Download Study<
⦿ DNA polymerase: klenTaq1 Elongation dynamics probed by Surface scanning smFRET

Förster resonance energy transfer (FRET) is an important biophysical method that provides unique insights into structural dynamics of biomolecular systems. Confocal FRET uses high spatiotemporal resolution (ns and 0.2 nm, respectivley) to investigate structural kinetics and intermediate states at single-molecule level. Fast molecule diffusion, however, limits the observation time. To surpass this limit, we development a technique that combines surface-scanning and covalent biomolecule immobilization on surfaces. This approach permits measurements over larger time scales, allowing the observation of low frequent structural exchanges, as shown for nucleotide addition of thermophilic bacterial Klentaq1 DNA polymerase. The high-throughput capability of this method yielded robust statistics and exhibited an average nucleotide addition rate of 12 nt/s. We identified that the Klentaq1 adopts three conformational states: an open state, an intermediate nt-binding state, and a closed state. In presence of an incorrect dNTP, we observed a slow process occurring between the nucleotide-binding and the open state. In contrast, in presence of a correct dNTP, the transition was fast and led to the closed state that allows catalysis.
⦿ gene therapy: Non-viral gene delivery vectors based on dynein fragment fusion proteins

Low efficiency of gene transfer is a recurrent problem in vaccine and gene therapy development using non-viral plasmid DNA (pDNA) vectors. During their transport to the target cell’s nuclei, plasmid vectors must overcome a series of physical, enzymatic, and diffusional barriers. In this work we developed a recombinant protein designed for pDNA delivery, transported from the cell periphery to the centrosome of mammalian cells, taking advantage of the molecular motors dynein. By fusing a DNA binding sequence to the N-terminus of the recombinant human dynein light chain LC8, the protein is able to interact and condense pDNA, generating positively charged complexes. Transfection of cultured HeLa cells confirmed the ability of the LC8 to facilitate pDNA uptake and indicate the involvement of the retrograde transport in the intracellular trafficking of pDNA:LC8 complexes via dynein. We further demonstrated a very low toxicity of the fusion protein vector, indicating great potential for in vivo applications. The newly developed modular shuttle protein exploit strategies used by viruses to infect mammalian cells to provide new approaches for gene therapy. >Download 2 Studies<
⦿ Model-free estimation of single DNA-protein interaction dissociation rates

Atomic Force Spectroscopy allows testing the free energy landscapes of biomolecular interactions. Usually, the dependency of the most probable rupture force on the force rate of the rupture force histogram is fitted with different approximative models. Here, we present a simple model free approach to extract the force-dependent dissociation rates directly from the force curve data. Simulations verified that dissociation rates at any force are directly reflected by the ratio of the number of detected rupture events. To calculate these dwell times of acting forces, all force curve data points of all curves measured can be taken into account, which significantly increases the amount of information which is considered for data analysis compared to other common methods used. Moreover, by providing force-dependent dissociation rates, this methodological approach allows direct testing and validating of any assumed energy landscape model. >Download Study<